How to merge SimpleCov results with parallel Rails specs on Semaphore CI
If you are running Rails specs on parallel machines with Knapsack Pro, one challenge you will run into is combining the code coverage results generated by SimpleCov.

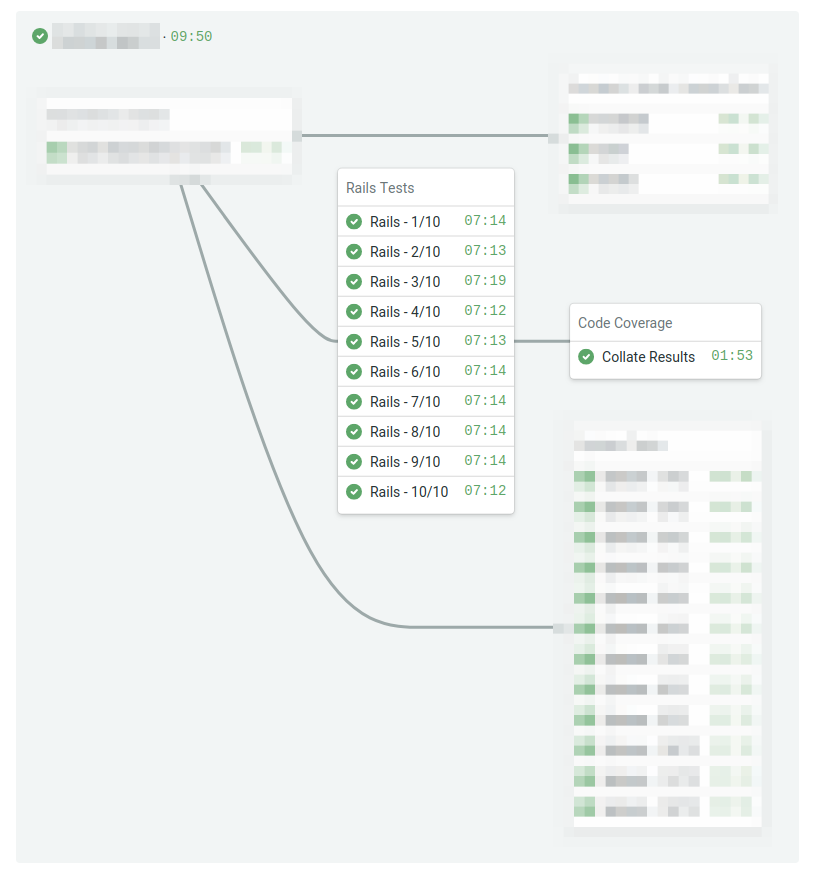
This post will show you how to generate a report of the total combined code coverage after all the tests have executed. Here’s what the pipeline diagram looks like at a high level with unrelated sections blurred out:
Configure
SimpleCov
First lets look at the config file used for SimpleCov below. Note no minimum_coverage configuration for failing the build. This is because each node most likely will not meet the minimum coverage threshold on its own so it could lead to the build failing erroneously.
Also note, before_queue hook for Knapsack Pro. This is the important piece, it will set a command name based on the CI node index so that the results are recorded against it.
.simplecov:
SimpleCov.start do
add_filter %r{^/config/}
add_filter %r{^/db/}
add_filter %r{^/spec/}
add_group 'Admin', 'app/admin'
add_group 'Controllers', 'app/controllers'
add_group 'Helpers', 'app/helpers'
add_group 'Jobs', 'app/jobs'
add_group 'Libraries', 'lib/'
add_group 'Mailers', 'app/mailers'
add_group 'Models', 'app/models'
add_group 'Policies', 'app/policies'
add_group 'Serializers', 'app/serializers'
end
Rails.application.eager_load!
KnapsackPro::Hooks::Queue.before_queue do
SimpleCov.command_name("rspec_ci_node_#{KnapsackPro::Config::Env.ci_node_index}")
endSo now when SimpleCov creates a .resultset.json it will have a specific key depending on which CI node it was run in like the example below. This will be useful down the line when it comes to combining the results.
{
"rspec_ci_node_0": {
"coverage": { ... }
}
}
{
"rspec_ci_node_1": {
"coverage": { ... }
}
}
...Semaphore CI
Below is the relevant portions of the Semaphore CI configuration. It runs the Rails tests and then uploads the coverage results as a Semaphore workflow artifact. After all the parallel tests have completed, it will run a job to collate the coverage results from all the machines.
.semaphore/semaphore.yml:
- name: "Rails Tests"
task:
jobs:
- name: Rails
parallelism: 10
commands:
- ./.semaphore/helpers/rails_tests.sh
epilogue:
always:
commands:
- ./.semaphore/helpers/upload_test_artifacts.sh
secrets:
- name: docker-hub
- name: knapsack-pro-rails
- name: "Code Coverage"
dependencies: ["Rails Tests"]
task:
env_vars:
- name: SEMAPHORE_RAILS_JOB_COUNT
value: "10"
jobs:
- name: Collate Results
commands:
- ./.semaphore/helpers/calc_code_coverage.sh
secrets:
- name: docker-hubBelow is the bash file which executes the Rails tests on each parallel machine. It sets up the Rails environment and then runs Knapsack Pro in Queue Mode.
.semaphore/helpers/rails_tests.sh:
#!/bin/bash
set -euo pipefail
docker-compose -f docker-compose.semaphore.yml --no-ansi run \
-e KNAPSACK_PRO_RSPEC_SPLIT_BY_TEST_EXAMPLES=true \
-e KNAPSACK_PRO_TEST_FILE_PATTERN="spec/**/*_spec.rb" \
ci bash -c "bin/rake ci:setup db:create db:structure:load knapsack_pro:queue:rspec['--no-color --format progress --format RspecJunitFormatter --out tmp/rspec-junit/rspec.xml']"If you want you can also learn how to use RspecJunitFormatter with KnapsackPro Queue Mode.
This is the bash file which is responsible for uploading the SimpleCov results from each machine. It compresses the coverage directory and uploads it to Semaphore.
.semaphore/helpers/upload_test_artifacts.sh:
if [ -d "tmp/rspec-junit" ]
then
echo "Pushing rspec junit results"
artifact push job tmp/rspec-junit --destination semaphore/test-results/
fi
if [ -d "coverage" ]
then
echo "Pushing simplecov results"
tar czf coverage_${SEMAPHORE_JOB_INDEX}.tgz -C coverage .
artifact push workflow coverage_${SEMAPHORE_JOB_INDEX}.tgz
fiLastly, this is the bash file for collating all the results from Semaphore. It will download the coverage artifacts from each parallel machine and run a rake task which will collate them and then upload the results into a combined total_coverage.tgz file as shown below:
.semaphore/helpers/calc_code_coverage.sh:
#!/bin/bash
set -euo pipefail
for i in $(eval echo "{1..$SEMAPHORE_RAILS_JOB_COUNT}"); do
artifact pull workflow coverage_$i.tgz;
mkdir coverage_$i
tar -xzf coverage_$i.tgz -C coverage_$i
done
docker-compose -f docker-compose.semaphore.yml --no-ansi run ci bash -c "bin/rake coverage:report"
tar czf total_coverage.tgz -C coverage .
artifact push workflow total_coverage.tgzThis coverage:report rake task will simply call SimpleCov.collate which will go through the coverage results in each folder and combine them into a single .resultset.json shown below
lib/task/coverage_report.rake:
namespace :coverage do
desc 'Collates all result sets generated by the different test runners'
task report: :environment do
require 'simplecov'
SimpleCov.collate Dir['coverage_*/.resultset.json']
end
end.resultset.json:
{
"rspec_ci_node_0, rspec_ci_node_1, rspec_ci_node_2, rspec_ci_node_3, rspec_ci_node_4, rspec_ci_node_5, rspec_ci_node_6, rspec_ci_node_7, rspec_ci_node_8, rspec_ci_node_9": {
"coverage": { ... }
}Summary
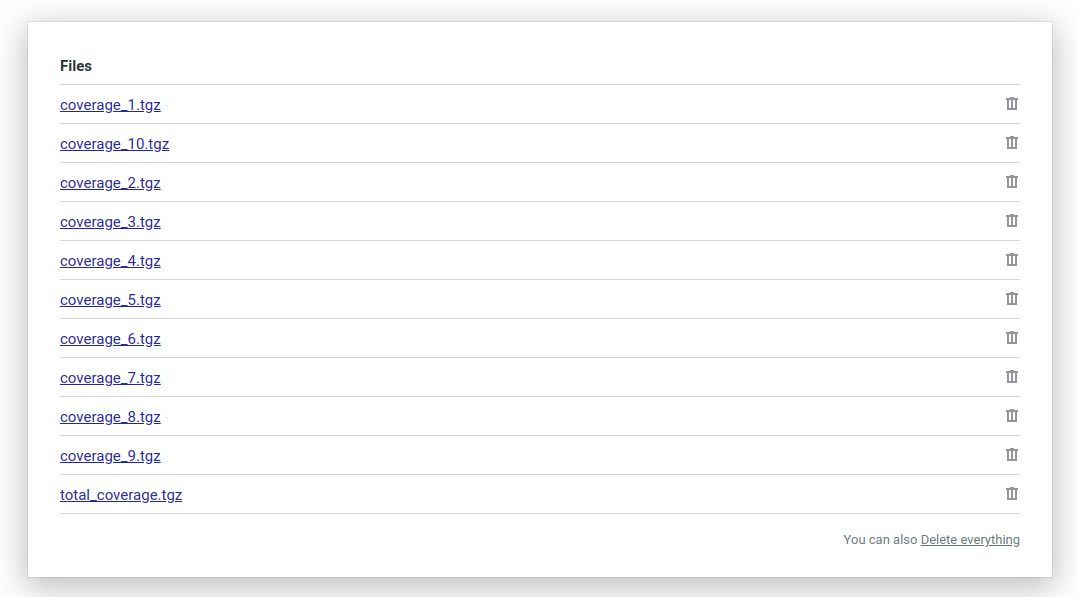
Finally here’s what your Semaphore workflow artifacts will look like. It will have a compressed coverage file generated on each machine and a total coverage file that we created at the very end:
This approach can also be easily ported over to other CI providers by simply changing the artifact push and artifact pull commands to S3 or another CI specific artifact upload command.
I hope this article was useful to you. Let me know if you have any questions or feedback. If you liked this post, please share it on social media and follow me on Twitter!